4 Statistics
We often summarize distributions with statistics: functions of data. The most basic way to do this is with summary, whose values can all be calculated individually. (E.g., the “mean” computes the [sum of all values] divided by [number of values].) There are many other statistics.
Code
4.1 Mean and Variance
The most basic statistics summarize the center of a distribution and how far apart the values are spread.
Mean.
Perhaps the most common statistic is the mean; \[\overline{X}=\frac{\sum_{i=1}^{N}X_{i}}{N},\] where \(X_{i}\) denotes the value of the \(i\)th observation.
Code
Variance.
Perhaps the second most common statistic is the variance: the average squared deviation from the mean \[V_{X} =\frac{\sum_{i=1}^{N} [X_{i} - \overline{X}]^2}{N}.\] The standard deviation is simply \(s_{X} = \sqrt{V_{X}}\).
Code

Note that a “corrected version” is used by R and many statisticians: \(V_{X} =\frac{\sum_{i=1}^{N} [X_{i} - \overline{X}]^2}{N-1}\).
Together, these statistics summarize the central tendency and dispersion of a distribution. In some special cases, such as with the normal distribution, they completely describe the distribution. Other distributions are easier to describe with other statistics.
4.2 Other Center/Spread Statistics
Absolute Deviations.
We can use the Median as a “robust alternative” to means. Recall that the \(q\)th quantile is the value where \(q\) percent of the data are below and (\(1-q\)) percent are above. The median (\(q=.5\)) is the point where half of the data is lower values and the other half is higher.
We can also use the Interquartile Range or Median Absolute Deviation as an alternative to variance. The first and third quartiles (\(q=.25\) and \(q=.75\)) together measure is the middle 50 percent of the data. The size of that range (interquartile range: the difference between the quartiles) represents “spread” or “dispersion” of the data. The median absolute deviation also measures spread \[ \tilde{X} = Med(X_{i}) \\ MAD_{X} = Med\left( | X_{i} - \tilde{X} | \right). \]
Code

Code
Note that there other absolute deviations:
4.3 Shape Statistics
Central tendency and dispersion are often insufficient to describe a distribution. To further describe shape, we can compute the “standard moments” skew and kurtosis, as well as other statistics.
Skewness.
This captures how symmetric the distribution is. \[Skew_{X} =\frac{\sum_{i=1}^{N} [X_{i} - \overline{X}]^3 / N}{ [s_{X}]^3 }\]
Kurtosis.
This captures how many “outliers” there are. \[Kurt_{X} =\frac{\sum_{i=1}^{N} [X_{i} - \overline{X}]^4 / N}{ [s_{X}]^4 }.\]
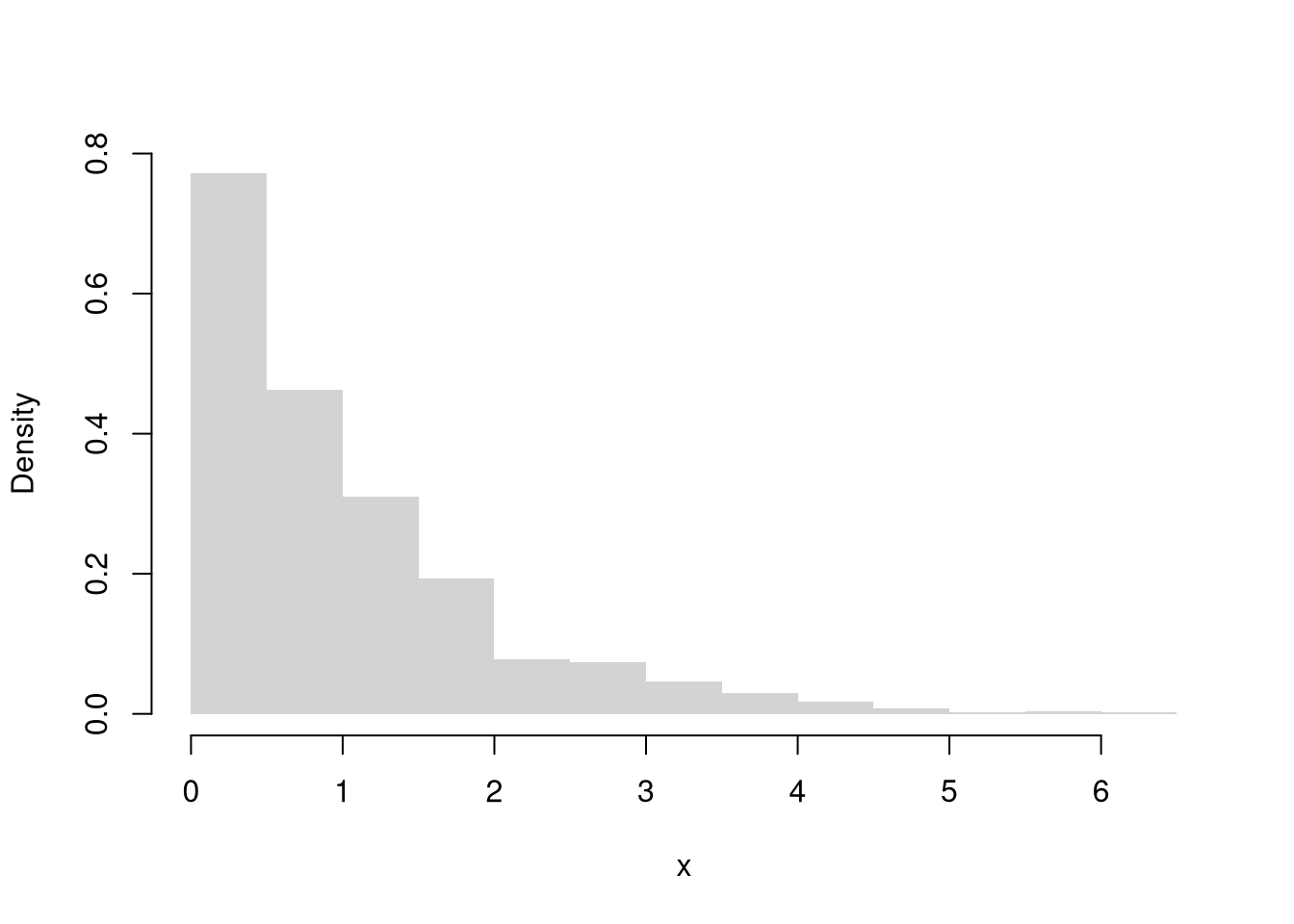
Clusters/Gaps.
You can also describe distributions in terms of how clustered the values are. Remember: a picture is worth a thousand words.
Code
# Random Number Generator
r_ugly1 <- function(n, theta1=c(-8,-1), theta2=c(-2,2), rho=.25){
omega <- rbinom(n, size=1, rho)
epsilon <- omega * runif(n, theta1[1], theta2[1]) +
(1-omega) * rnorm(n, theta1[2], theta2[2])
return(epsilon)
}
# Large Sample
par(mfrow=c(1,1))
X <- seq(-12,6,by=.001)
rx <- r_ugly1(1000000)
hist(rx, breaks=1000, freq=F, border=NA,
xlab="x", main='')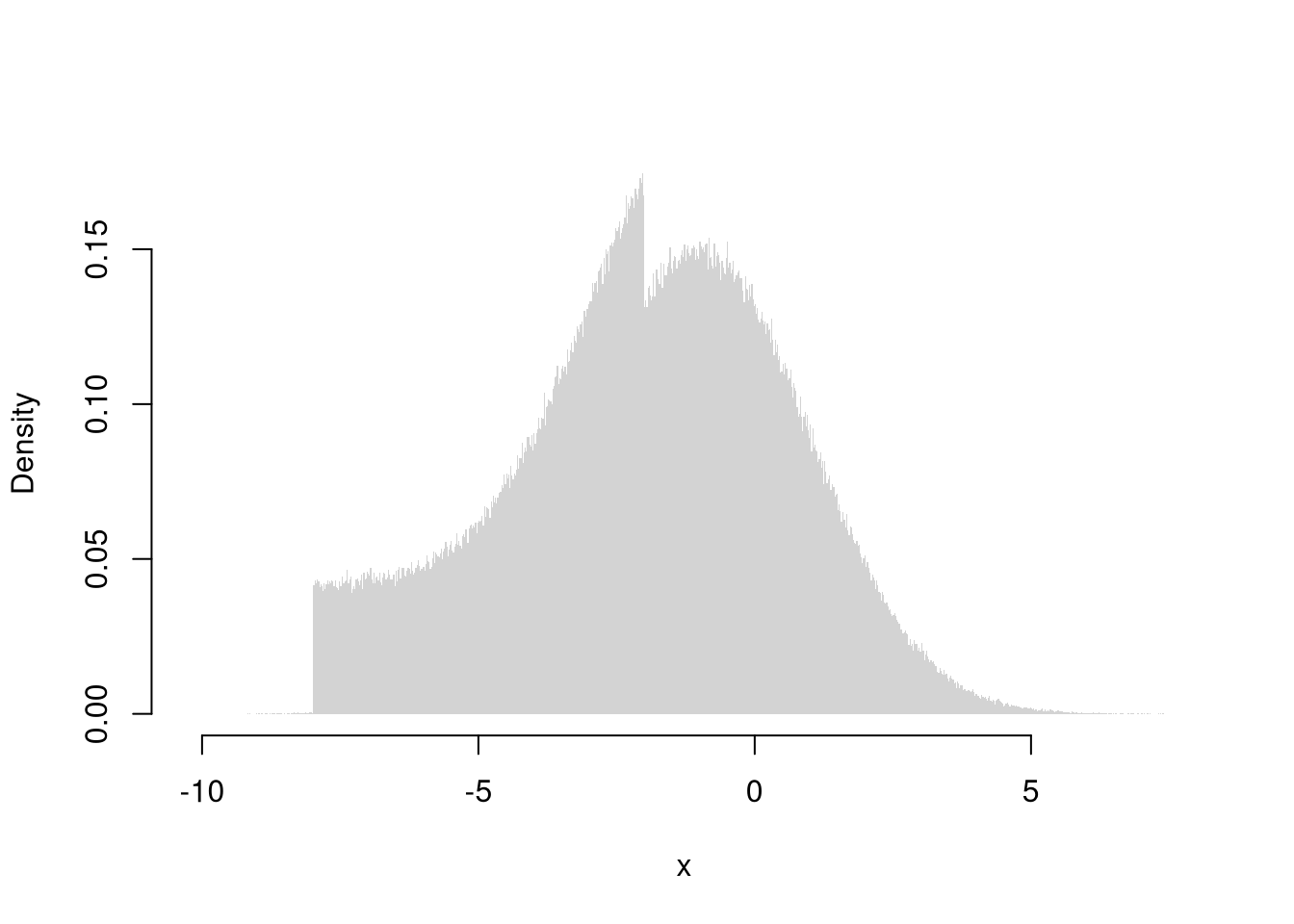
4.4 Associations
There are several ways to quantitatively describe the relationship between two variables, \(Y\) and \(X\). The major differences surround whether the variables are cardinal, ordinal, or categorical.
Correlation.
Pearson (Linear) Correlation. Suppose \(X\) and \(Y\) are both cardinal data. As such, you can compute the most famous measure of association, the covariance: \[ C_{XY} = \sum_{i} [X_i - \overline{X}] [Y_i - \overline{Y}] / N \]
Code
Note that \(C_{XX}=V_{X}\). For ease of interpretation, we rescale this statistic to always lay between \(-1\) and \(1\) \[ r_{XY} = \frac{ C_{XY} }{ \sqrt{V_X} \sqrt{V_Y}} \]
Falk Codeviance. The Codeviance is a robust alternative to Covariance. Instead of relying on means (which can be sensitive to outliers), it uses medians (\(\tilde{X}\)) to capture the central tendency.3 We can also scale the Codeviance by the median absolute deviation to compute the median correlation. \[ \text{CoDev}(X,Y) = \text{Med}\left\{ |X_i - \tilde{X}| |Y_i - \tilde{Y}| \right\} \\ \tilde{r}_{XY} = \frac{ \text{CoDev}(X,Y) }{ \text{MAD}(X) \text{MAD}(Y) }. \]
Code
cor_m <- function(xy) {
# Compute medians for each column
med <- apply(xy, 2, median)
# Subtract the medians from each column
xm <- sweep(xy, 2, med, "-")
# Compute CoDev
CoDev <- median(xm[, 1] * xm[, 2])
# Compute the medians of absolute deviation
MadProd <- prod( apply(abs(xm), 2, median) )
# Return the robust correlation measure
return( CoDev / MadProd)
}
cor_m(xy)
## [1] 0.005707763Kendall’s Tau.
Suppose \(X\) and \(Y\) are both ordered variables. Kendall’s Tau measures the strength and direction of association by counting the number of concordant pairs (where the ranks agree) versus discordant pairs (where the ranks disagree). A value of \(\tau = 1\) implies perfect agreement in rankings, \(\tau = -1\) indicates perfect disagreement, and \(\tau = 0\) suggests no association in the ordering. \[ \tau = \frac{2}{n(n-1)} \sum_{i} \sum_{j > i} \text{sgn} \Bigl( (X_i - X_j)(Y_i - Y_j) \Bigr), \] where the sign function is: \[ \text{sgn}(z) = \begin{cases} +1 & \text{if } z > 0\\ 0 & \text{if } z = 0 \\ -1 & \text{if} z < 0 \end{cases}. \]
Cramer’s V.
Suppose \(X\) and \(Y\) are both categorical variables; the value of \(X\) is one of \(1...r\) categories and the value of \(Y\) is one of \(1...k\) categories. Cramer’s V quantifies the strength of association by adjusting a “chi-squared” statistic to provide a measure that ranges from 0 to 1; 0 indicates no association while a value closer to 1 signifies a strong association.
First, consider a contingency table for \(X\) and \(Y\) with \(r\) rows and \(k\) columns. The chi-square statistic is then defined as:
\[ \chi^2 = \sum_{i=1}^{r} \sum_{j=1}^{k} \frac{(O_{ij} - E_{ij})^2}{E_{ij}}. \]
where
- \(O_{ij}\) denote the observed frequency in cell \((i, j)\),
- \(E_{ij} = \frac{R_i \cdot C_j}{n}\) is the expected frequency for each cell if \(X\) and \(Y\) are independent
- \(R_i\) denote the total frequency for row \(i\) (i.e., \(R_i = \sum_{j=1}^{k} O_{ij}\)),
- \(C_j\) denote the total frequency for column \(j\) (i.e., \(C_j = \sum_{i=1}^{r} O_{ij}\)),
- \(n\) be the grand total of observations, so that \(n = \sum_{i=1}^{r} \sum_{j=1}^{k} O_{ij}\).
Second, normalize the chi-square statistic with the sample size and the degrees of freedom to compute Cramer’s V.
\[ V = \sqrt{\frac{\chi^2 / n}{\min(k - 1, \, r - 1)}}, \]
where:
- \(n\) is the total sample size,
- \(k\) is the number of categories for one variable,
- \(r\) is the number of categories for the other variable.
Code
xy <- USArrests[,c('Murder','UrbanPop')]
xy[,1] <- cut(xy[,1],3)
xy[,2] <- cut(xy[,2],4)
table(xy)
## UrbanPop
## Murder (31.9,46.8] (46.8,61.5] (61.5,76.2] (76.2,91.1]
## (0.783,6.33] 4 5 8 5
## (6.33,11.9] 0 4 7 6
## (11.9,17.4] 2 4 2 3
cor_v <- function(xy){
# Create a contingency table from the categorical variables
tbl <- table(xy)
# Compute the chi-square statistic (without Yates' continuity correction)
chi2 <- chisq.test(tbl, correct=FALSE)$statistic
# Total sample size
n <- sum(tbl)
# Compute the minimum degrees of freedom (min(rows-1, columns-1))
df_min <- min(nrow(tbl) - 1, ncol(tbl) - 1)
# Calculate Cramer's V
V <- sqrt((chi2 / n) / df_min)
return(V)
}
cor_v(xy)
## X-squared
## 0.2307071
# DescTools::CramerV( table(xy) )4.5 Beyond Basics
Use expansion “packages” for less common procedures and more functionality
CRAN.
Most packages can be found on CRAN and can be easily installed
Code
The most common tasks also have cheatsheets you can use.
For example, to generate ‘exotic’ probability distributions
Code
Code
Github.
Sometimes you will want to install a package from GitHub. For this, you can use devtools or its light-weight version remotes
Note that to install devtools, you also need to have developer tools installed on your computer.
To color terminal output on Linux systems, you can use the colorout package
4.6 Further Reading
Many random variables are related to each other
- https://en.wikipedia.org/wiki/Relationships_among_probability_distributions
- https://www.math.wm.edu/~leemis/chart/UDR/UDR.html
- https://qiangbo-workspace.oss-cn-shanghai.aliyuncs.com/2018-11-11-common-probability-distributions/distab.pdf
Note that numbers randomly generated on your computer cannot be truly random, they are “Pseudorandom”.
See also Theil-Sen Estimator, which may be seen as a precursor.↩︎