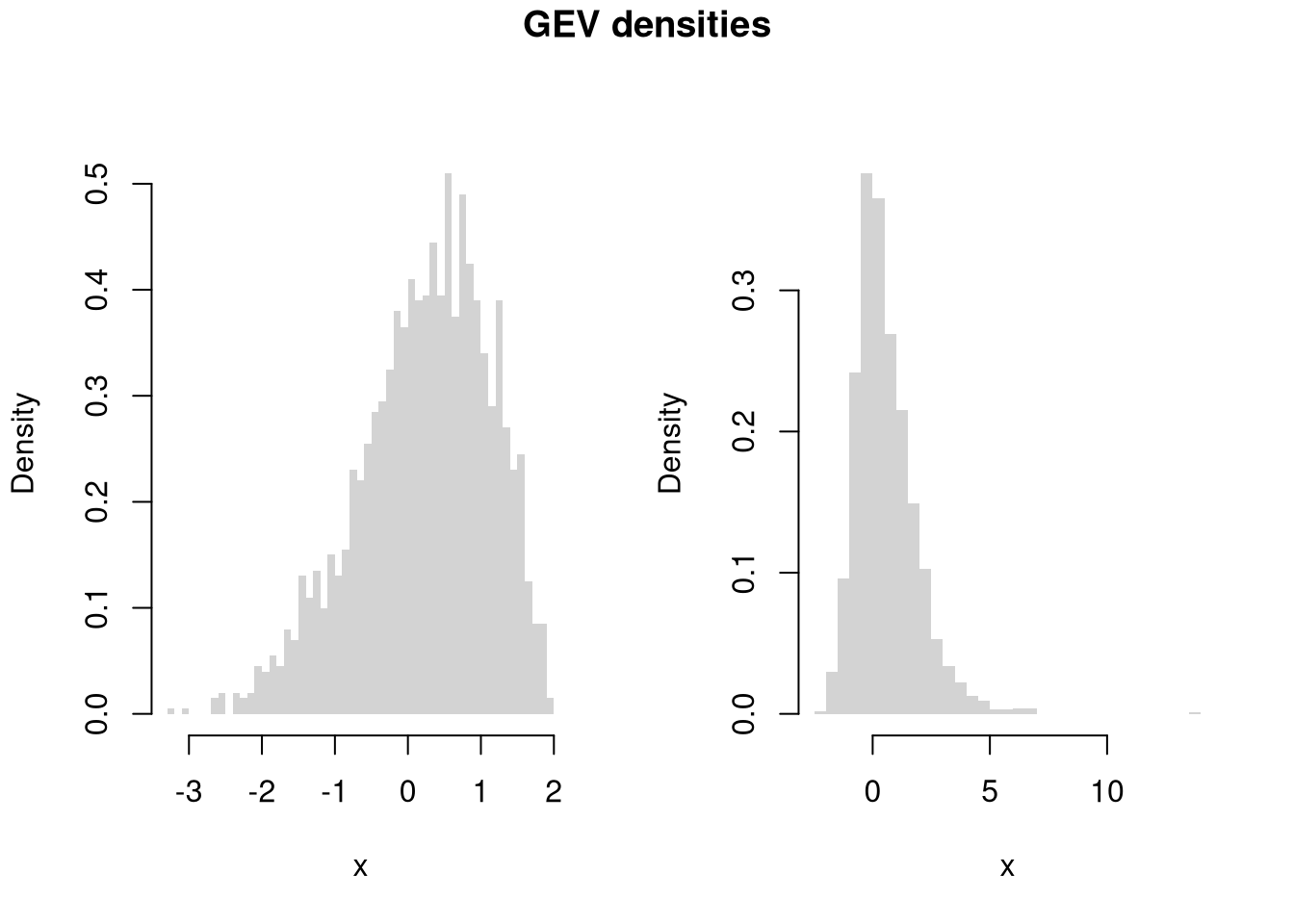
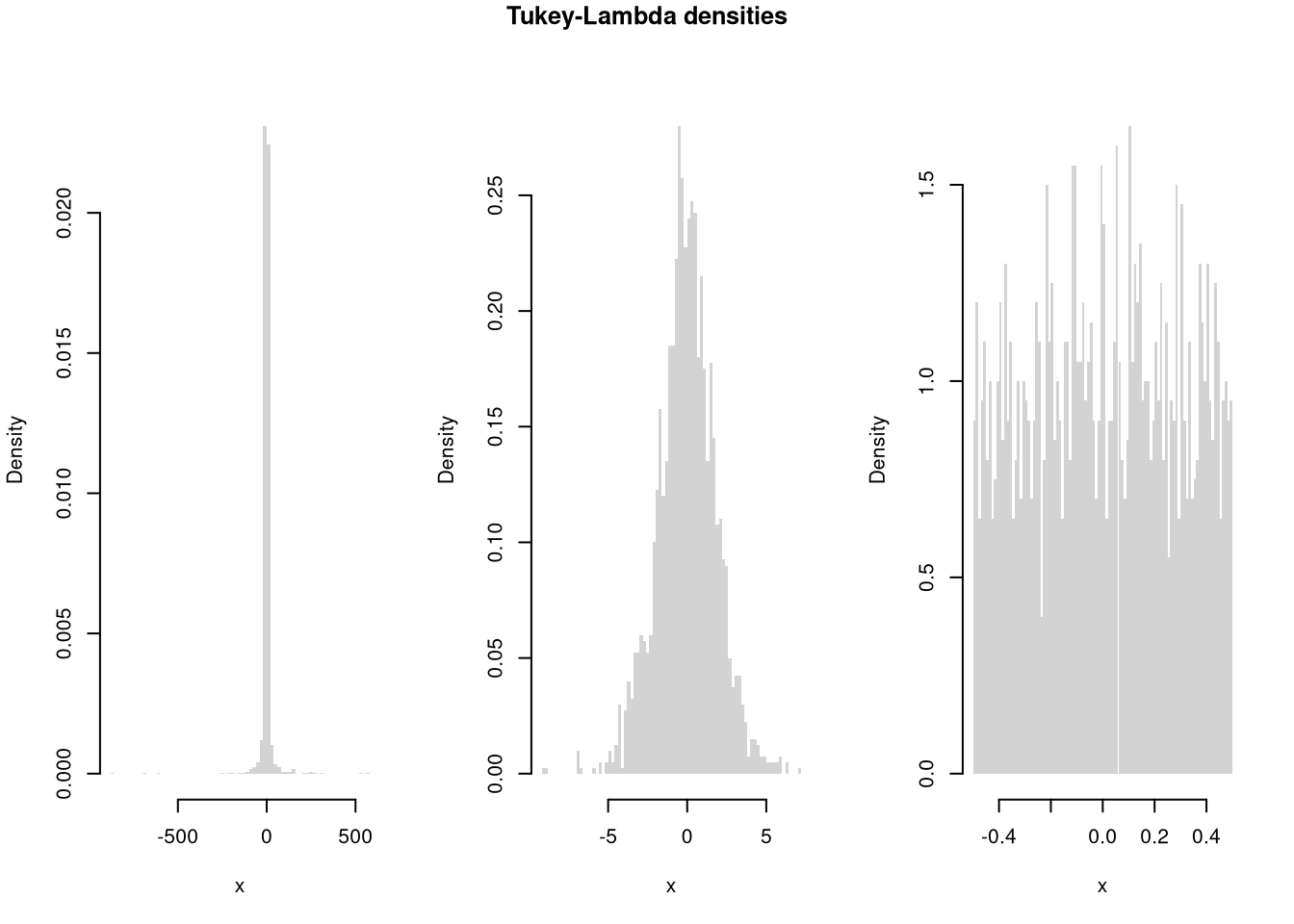
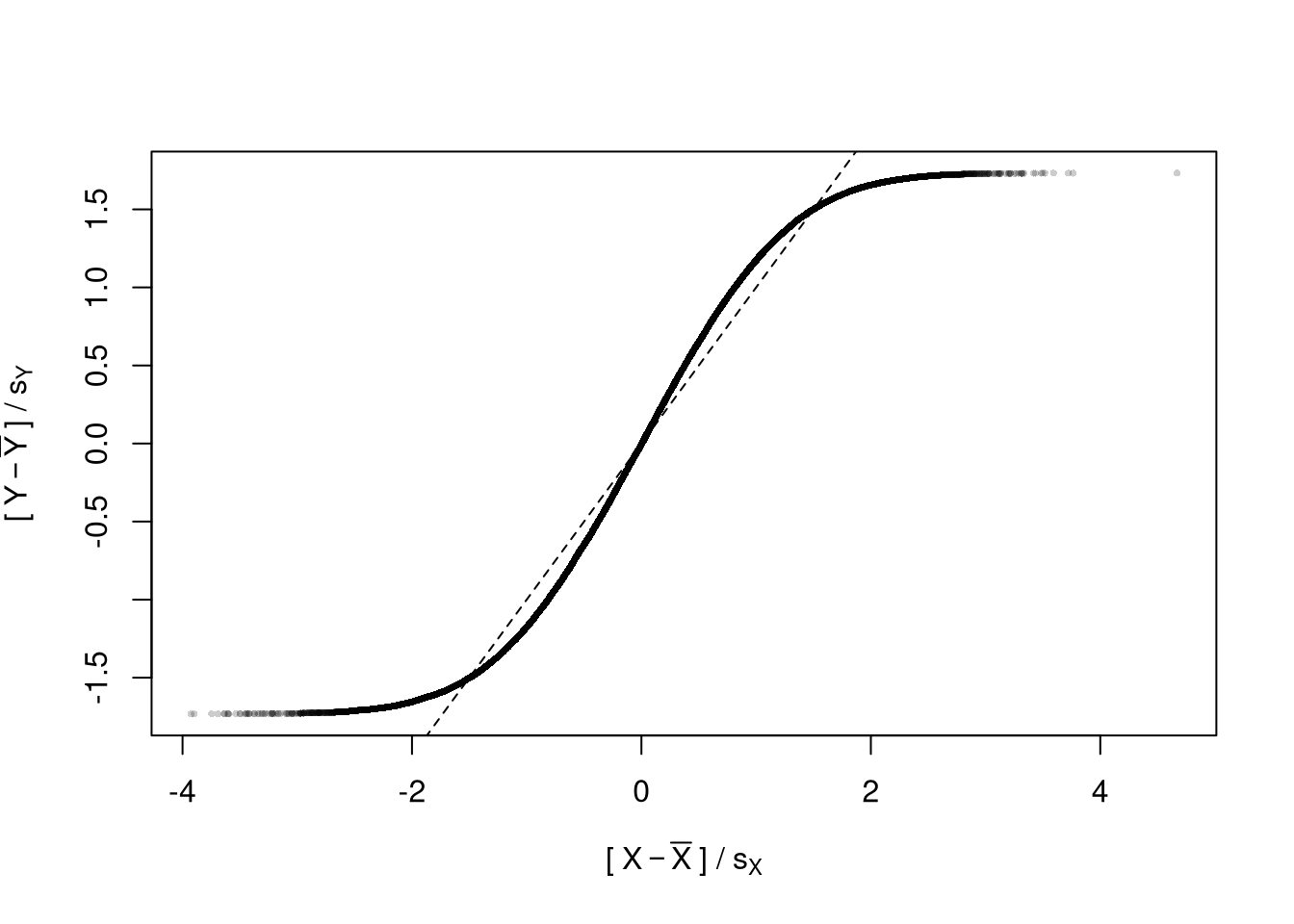
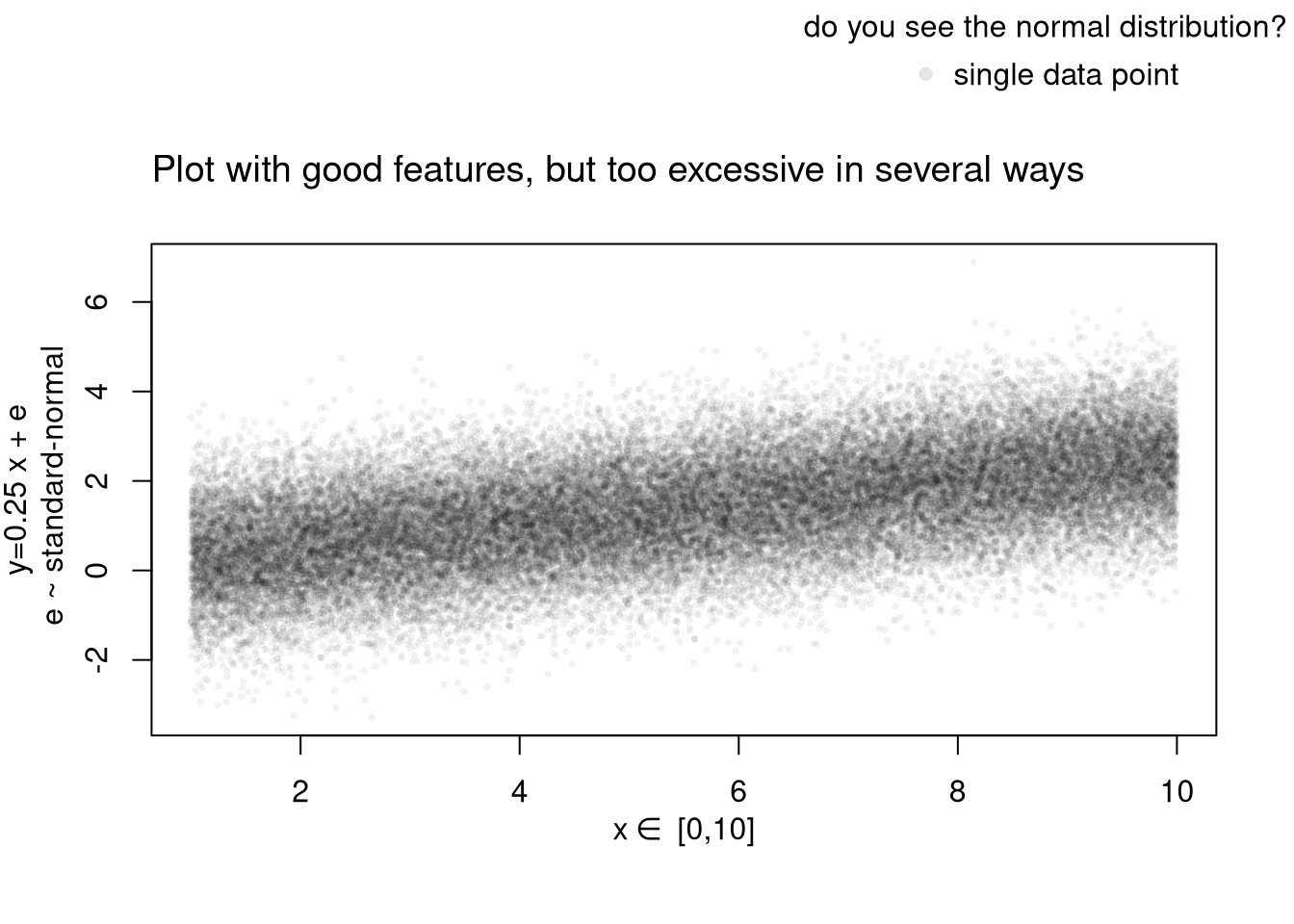
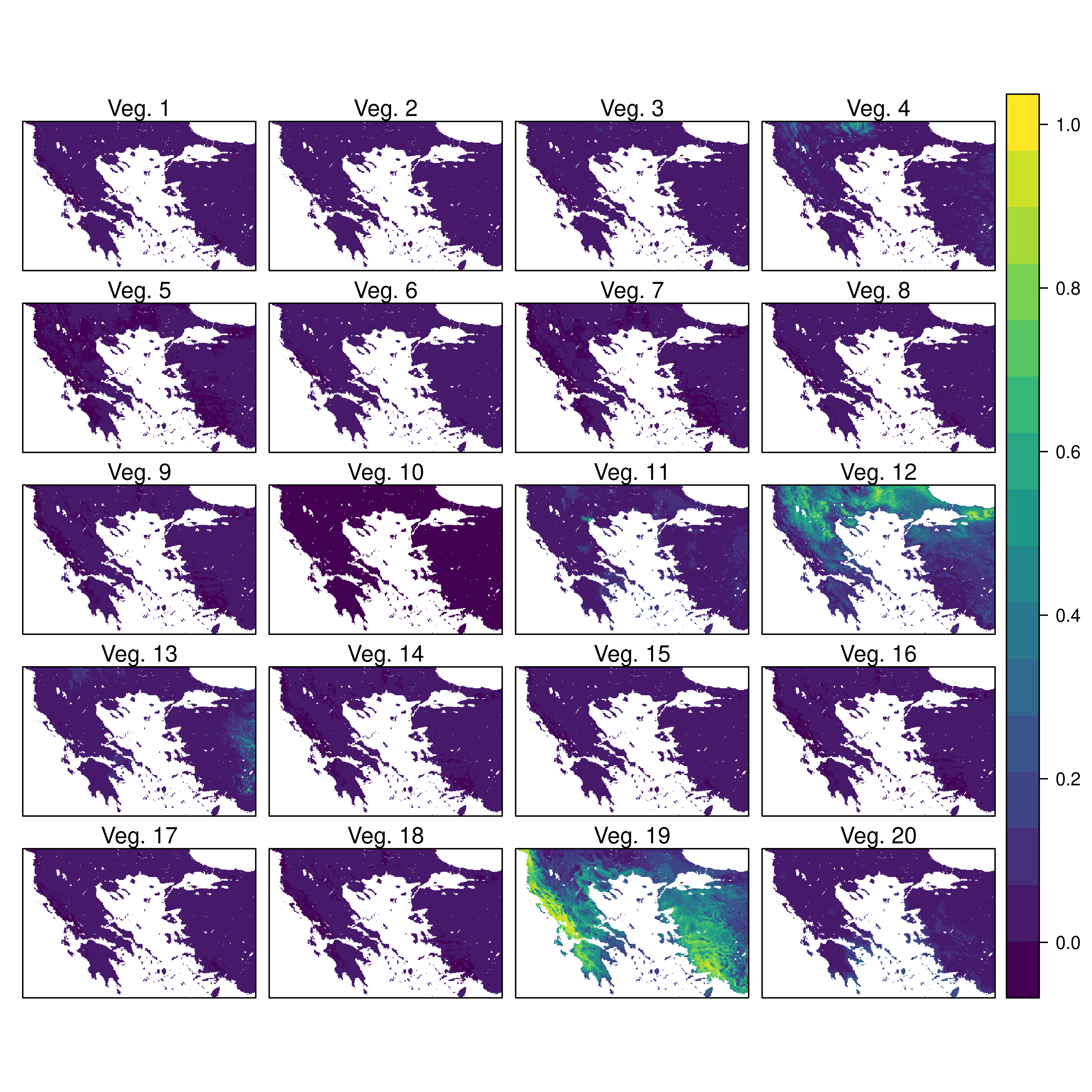
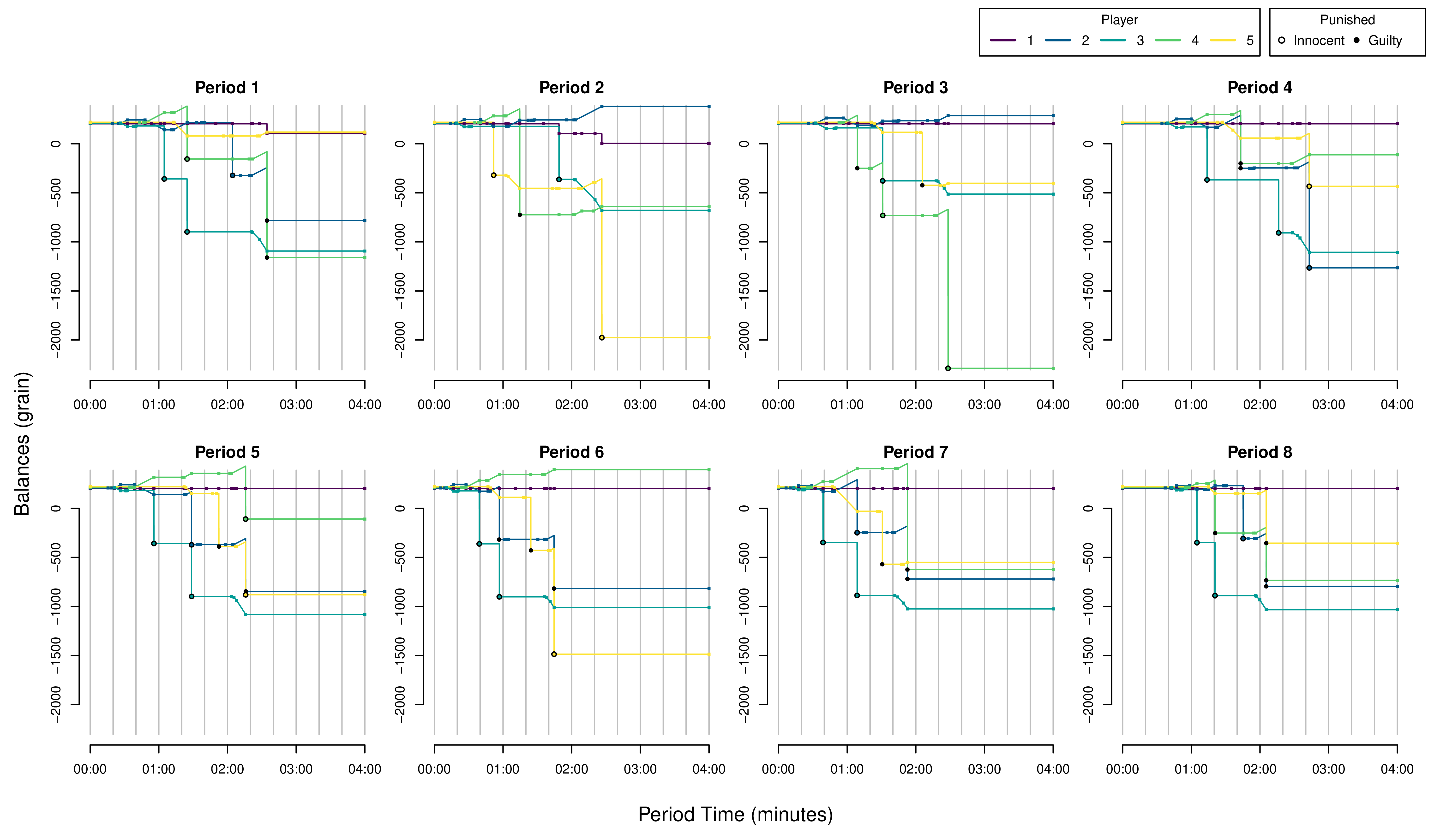
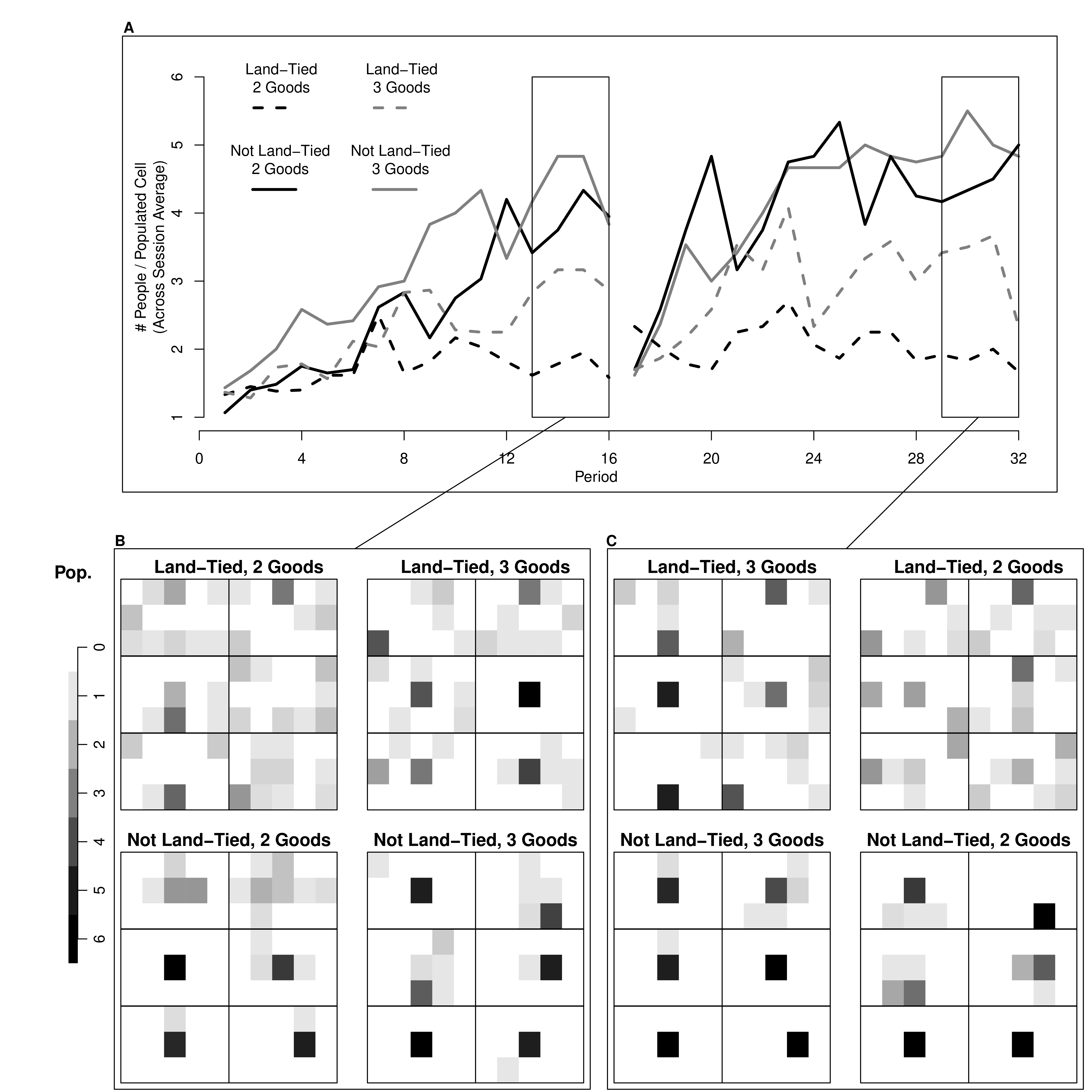
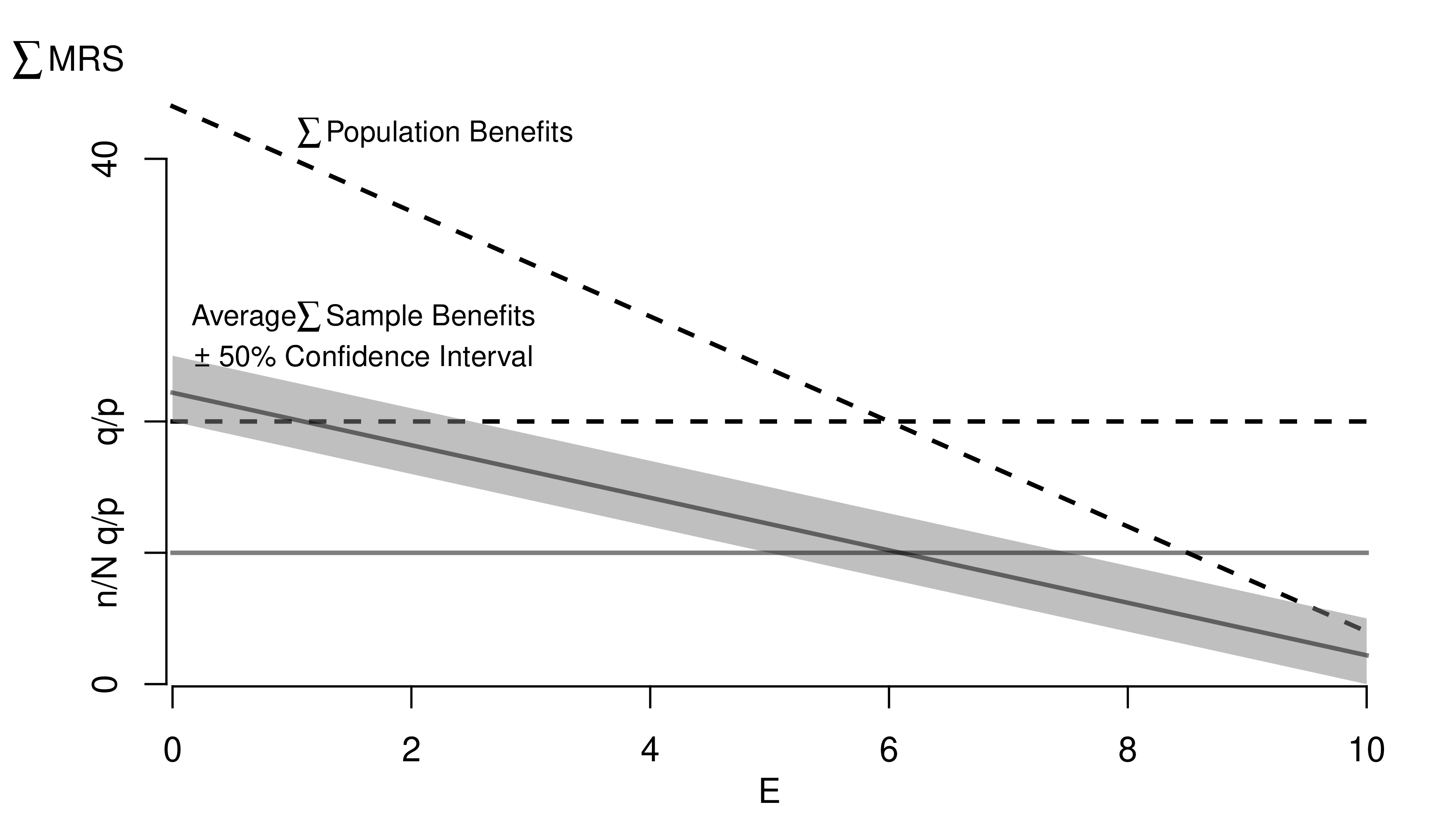
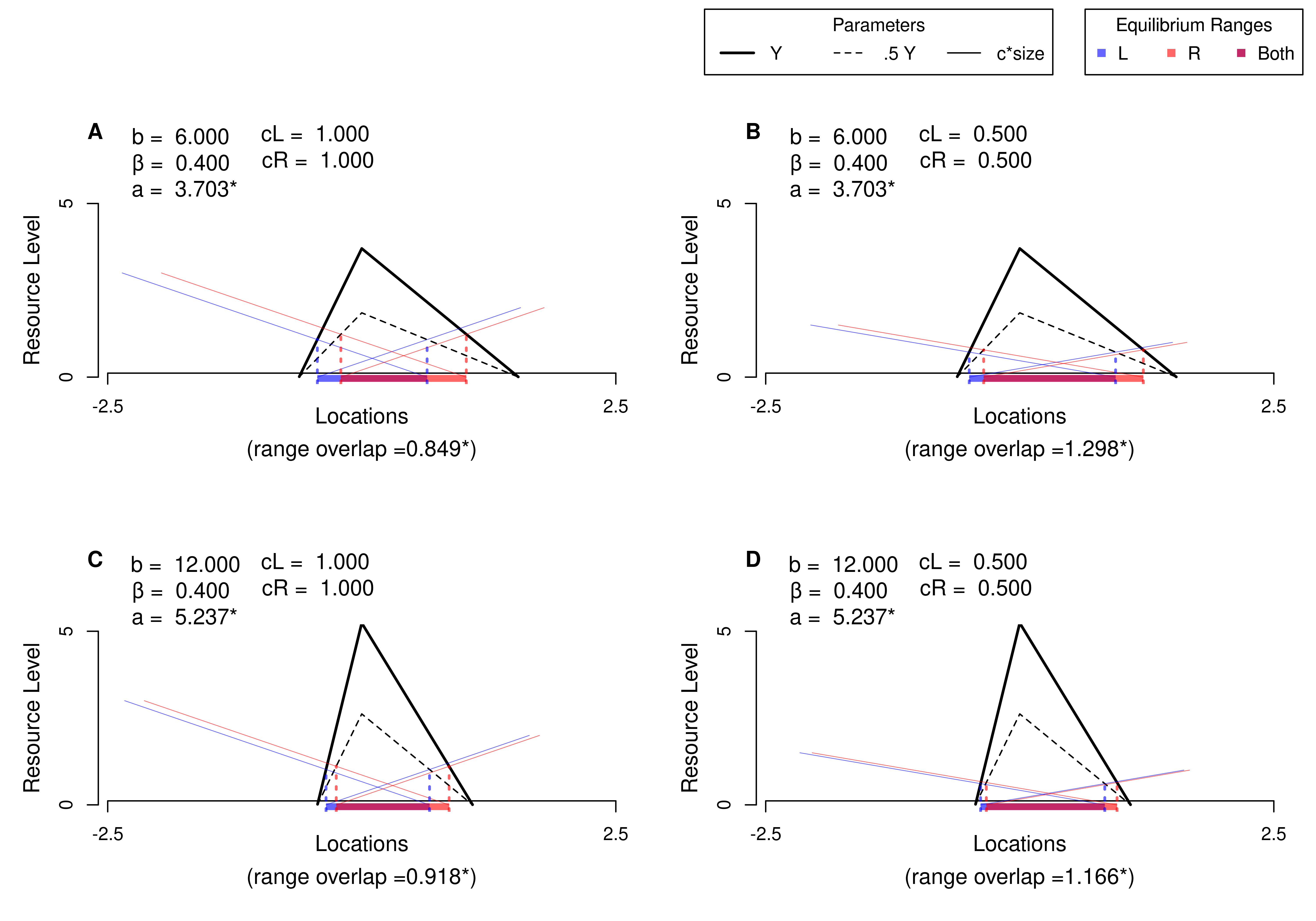
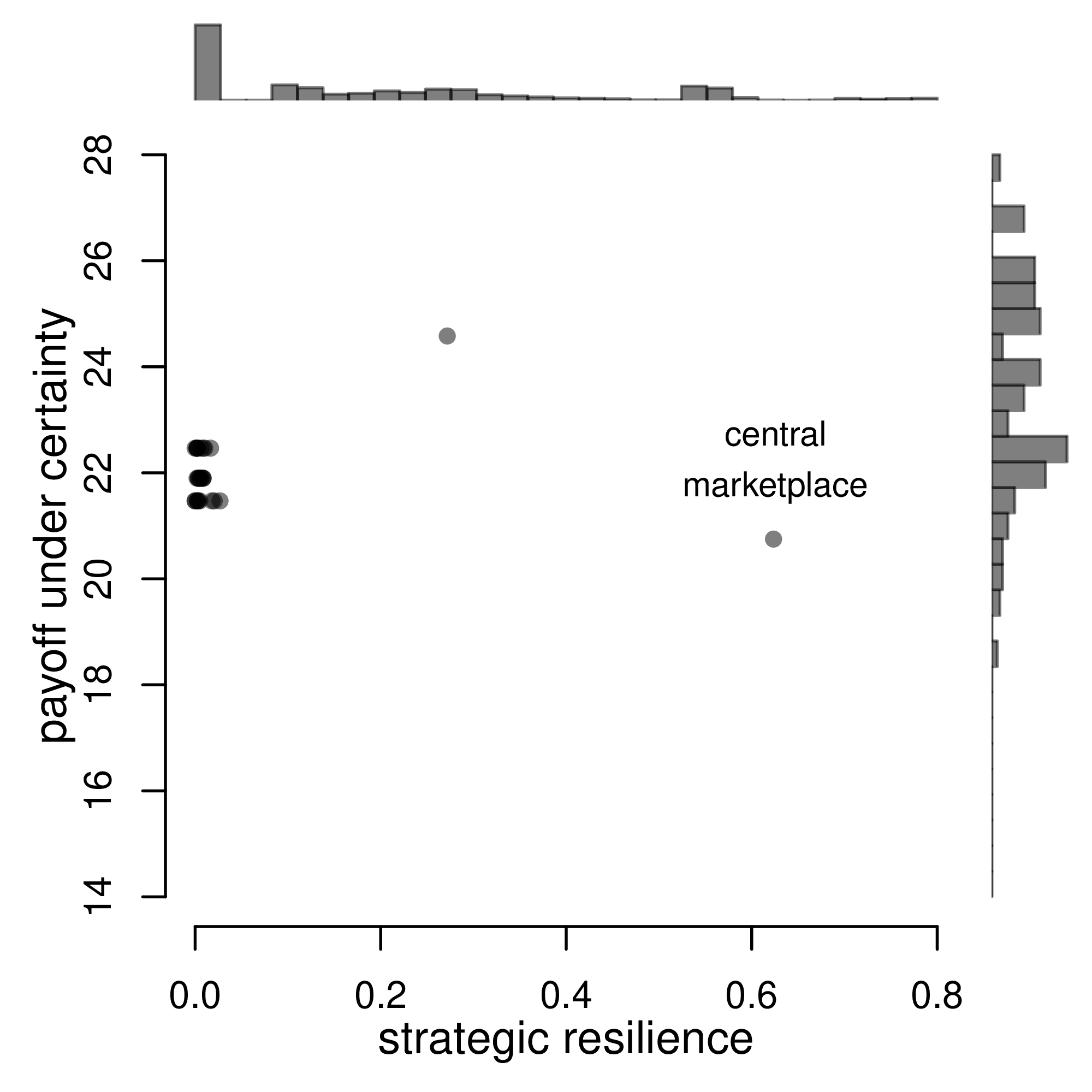
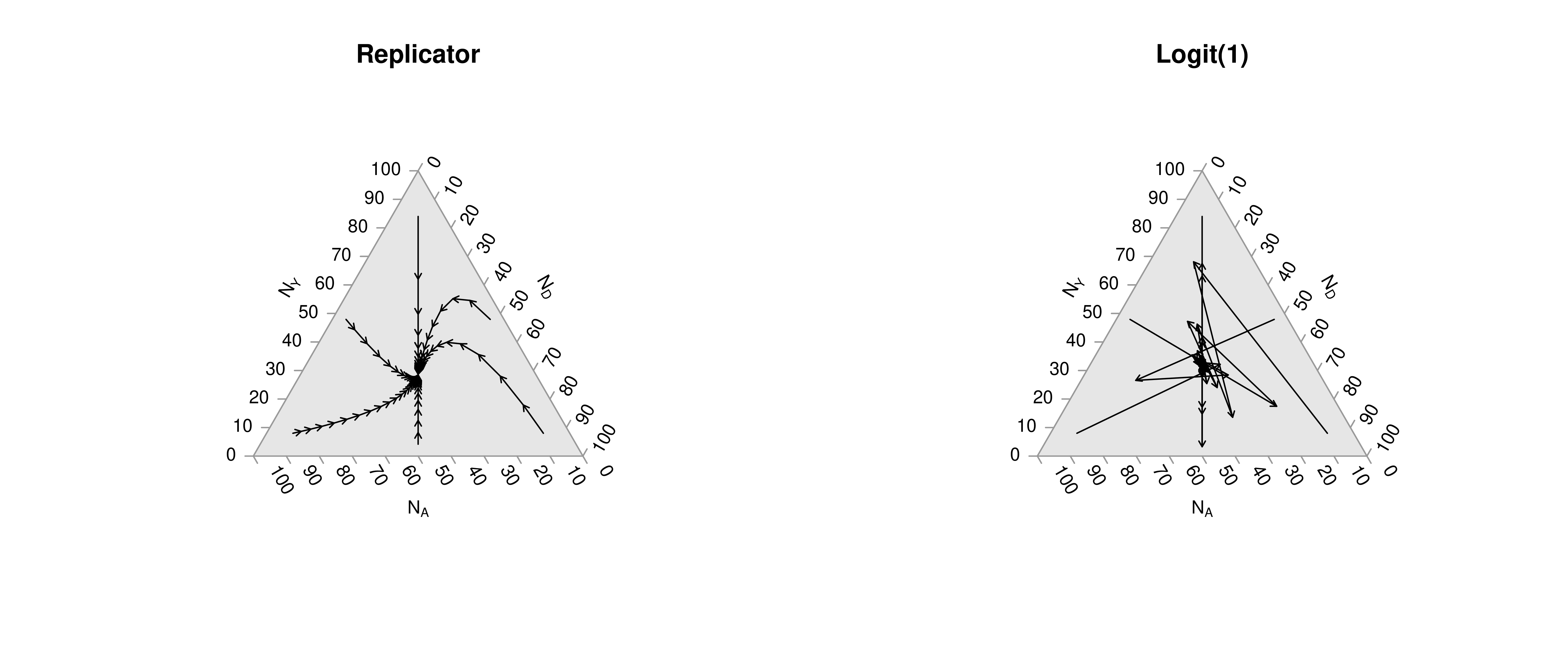
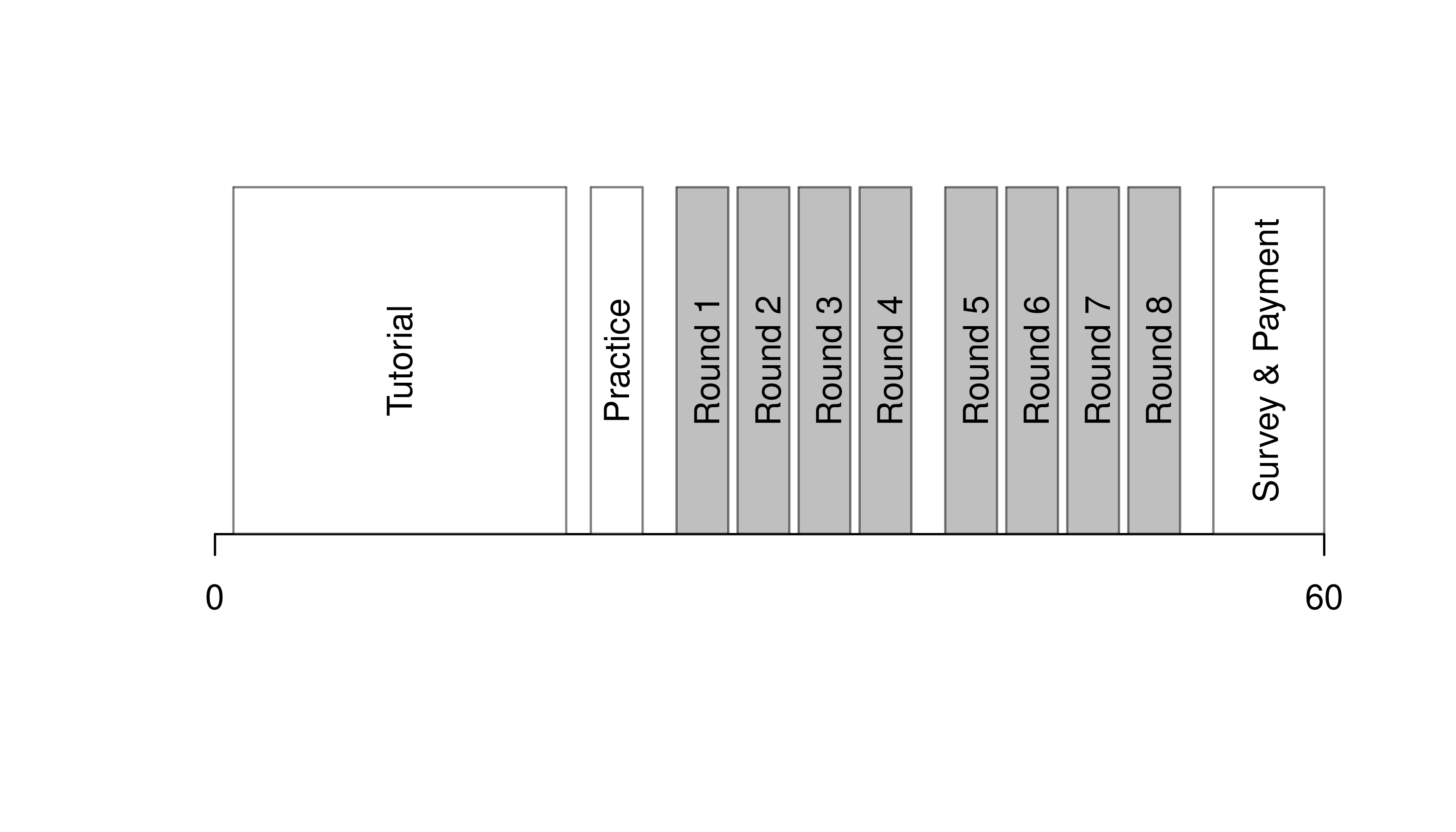
# Another Example
xy_dat <- data.frame(x=x, y=y)
par(fig=c(0,1,0,0.9), new=F)
plot(y~x, xy_dat, pch=16, col=rgb(0,0,0,.05), cex=.5,
xlab='', ylab='') # Format Axis Labels Seperately
mtext( 'y=0.25 x + e\n e ~ standard-normal', 2, line=2.2)
mtext( expression(x%in%~'[0,10]'), 1, line=2.2)
#abline( lm(y~x, data=xy_dat), lty=2)
title('Plot with good features, but too excessive in several ways',
adj=0, font.main=1)
# Outer Legend (https://stackoverflow.com/questions/3932038/)
outer_legend <- function(...) {
opar <- par(fig=c(0, 1, 0, 1), oma=c(0, 0, 0, 0),
mar=c(0, 0, 0, 0), new=TRUE)
on.exit(par(opar))
plot(0, 0, type='n', bty='n', xaxt='n', yaxt='n')
legend(...)
}
outer_legend('topright', legend='single data point',
title='do you see the normal distribution?',
pch=16, col=rgb(0,0,0,.1), cex=1, bty='n')