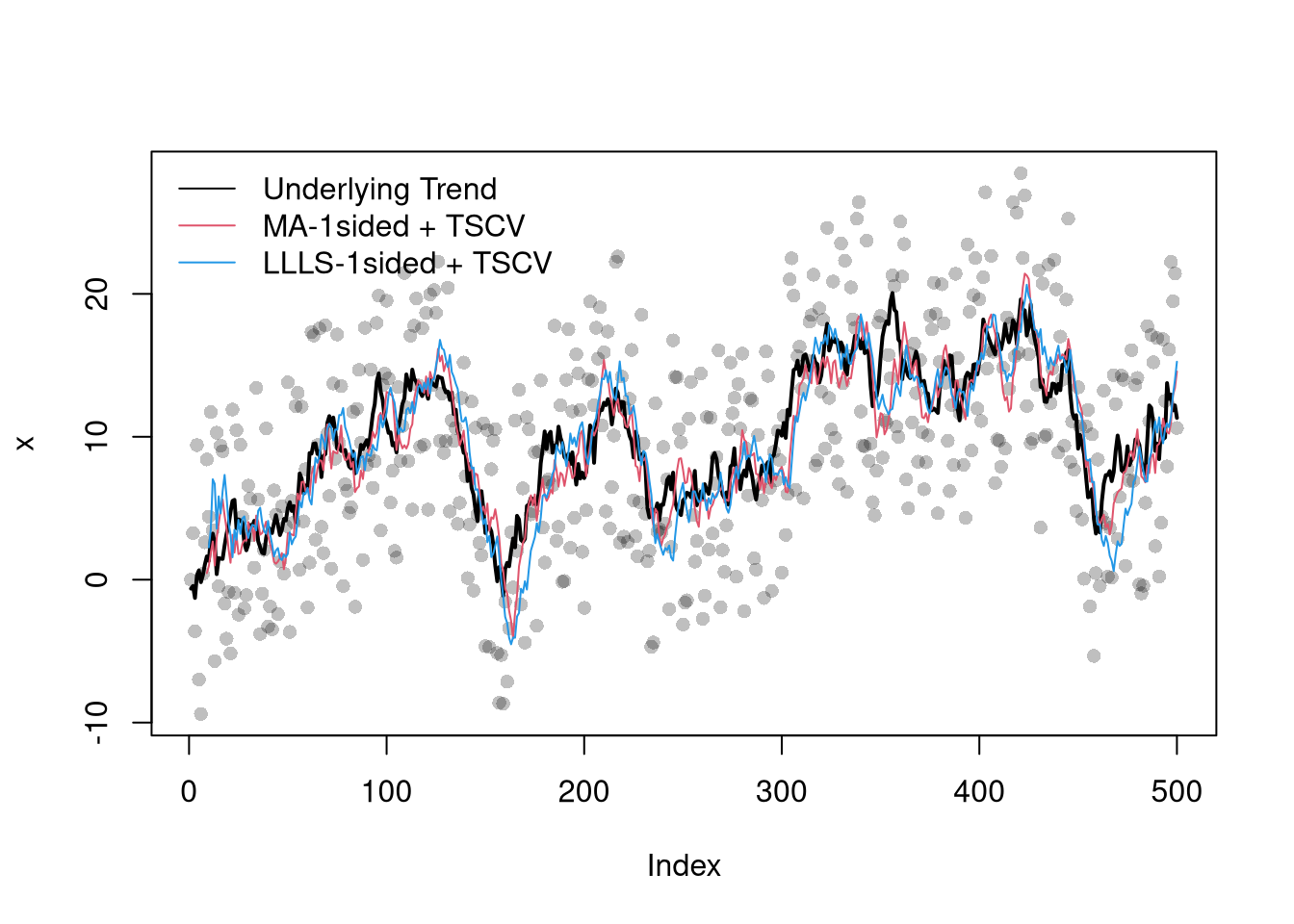
## Plot Simulated Data
x <- dat$x
x0 <- dat$x0
par(fig = c(0,1,0,1), new=F)
plot(x, pch=16, col=grey(0,.25))
lines(x0, col=1, lty=1, lwd=2)
## Work with differenced data?
#n <- length(Yt)
#plot(Yt[1:(n-1)], Yt[2:n],
# xlab='d Y (t-1)', ylab='d Y (t)',
# col=grey(0,.5), pch=16)
#Yt <- diff(Yt)
## TSCV One Sided Moving Average
filter_bws <- seq(1,20,by=1)
filter_mape_bws <- sapply(filter_bws, function(h){
bw <- c(0,rep(1/h,h)) ## Leave current one out
s2 <- filter(x, bw, sides=1)
pe <- s2 - x
mape <- mean( abs(pe)^2, na.rm=T)
})
filter_mape_star <- filter_mape_bws[which.min(filter_mape_bws)]
filter_h_star <- filter_bws[which.min(filter_mape_bws)]
filter_tscv <- filter(x, c(rep(1/filter_h_star,filter_h_star)), sides=1)
# Plot Optimization Results
#par(fig = c(0.07, 0.35, 0.07, 0.35), new=T)
#plot(filter_bws, filter_mape_bws, type='o', ylab='mape', pch=16)
#points(filter_h_star, filter_mape_star, pch=19, col=2, cex=1.5)
## TSCV for LLLS
library(np)
llls_bws <- seq(8,28,by=1)
llls_burnin <- 10
llls_mape_bws <- sapply(llls_bws, function(h){ # cat(h,'\n')
pe <- sapply(llls_burnin:nrow(dat), function(t_end){
dat_t <- dat[dat$t<t_end, ]
reg <- npreg(x~t, data=dat_t, bws=h,
ckertype='epanechnikov',
bandwidth.compute=F, regtype='ll')
edat <- dat[dat$t==t_end,]
pred <- predict(reg, newdata=edat)
pe <- edat$x - pred
return(pe)
})
mape <- mean( abs(pe)^2, na.rm=T)
})
llls_mape_star <- llls_mape_bws[which.min(llls_mape_bws)]
llls_h_star <- llls_bws[which.min(llls_mape_bws)]
#llls_tscv <- predict( npreg(x~t, data=dat, bws=llls_h_star,
# bandwidth.compute=F, regtype='ll', ckertype='epanechnikov'))
llls_tscv <- sapply(llls_burnin:nrow(dat), function(t_end, h=llls_h_star){
dat_t <- dat[dat$t<t_end, ]
reg <- npreg(x~t, data=dat_t, bws=h,
ckertype='epanechnikov',
bandwidth.compute=F, regtype='ll')
edat <- dat[dat$t==t_end,]
pred <- predict(reg, newdata=edat)
return(pred)
})
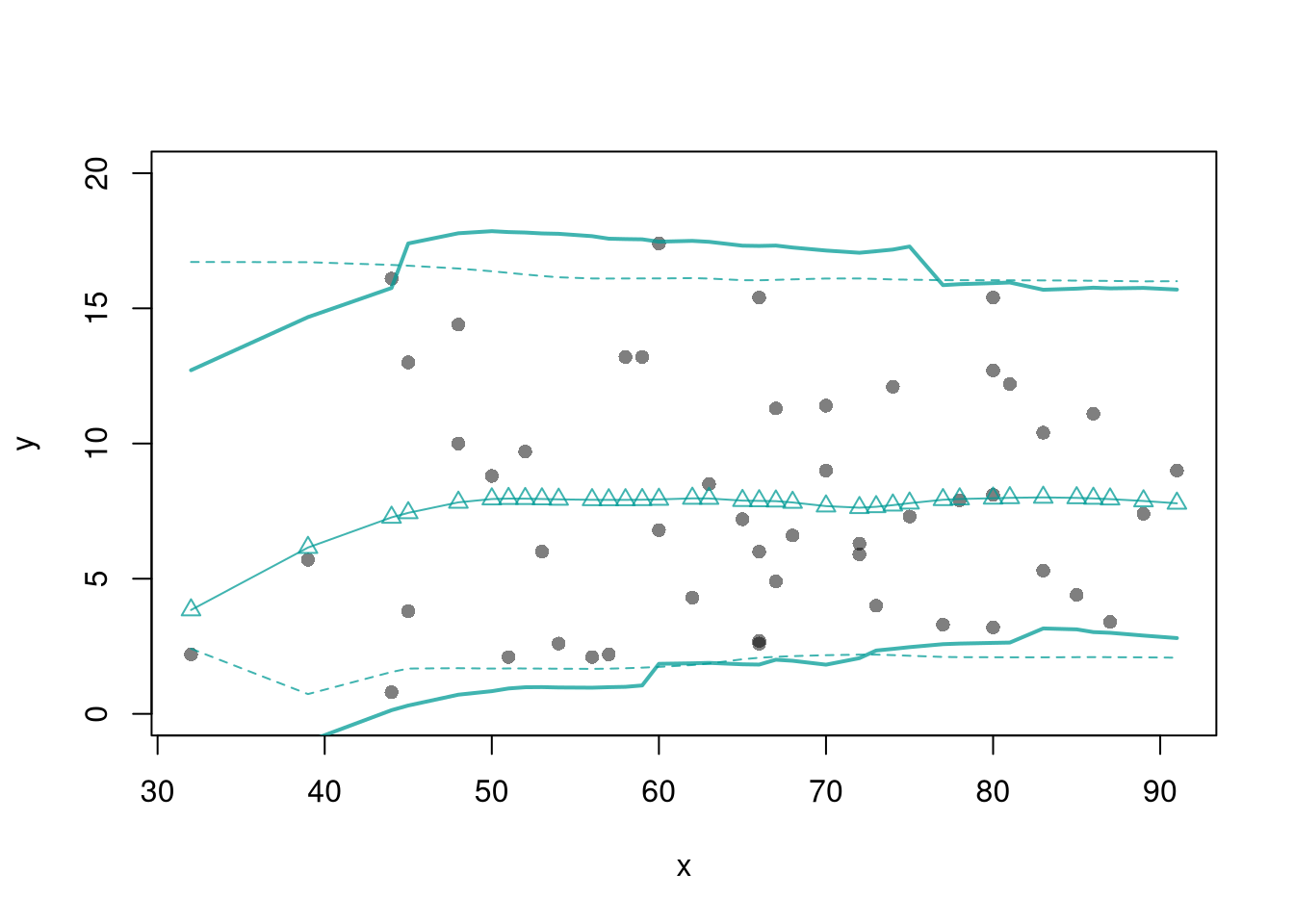
## Compare Fits Qualitatively
lines(filter_tscv, col=2, lty=1, lwd=1)
lines(llls_burnin:nrow(dat), llls_tscv, col=4, lty=1, lwd=1)
legend('topleft', lty=1, col=c(1,2,4), bty='n',
c('Underlying Trend', 'MA-1sided + TSCV', 'LLLS-1sided + TSCV'))