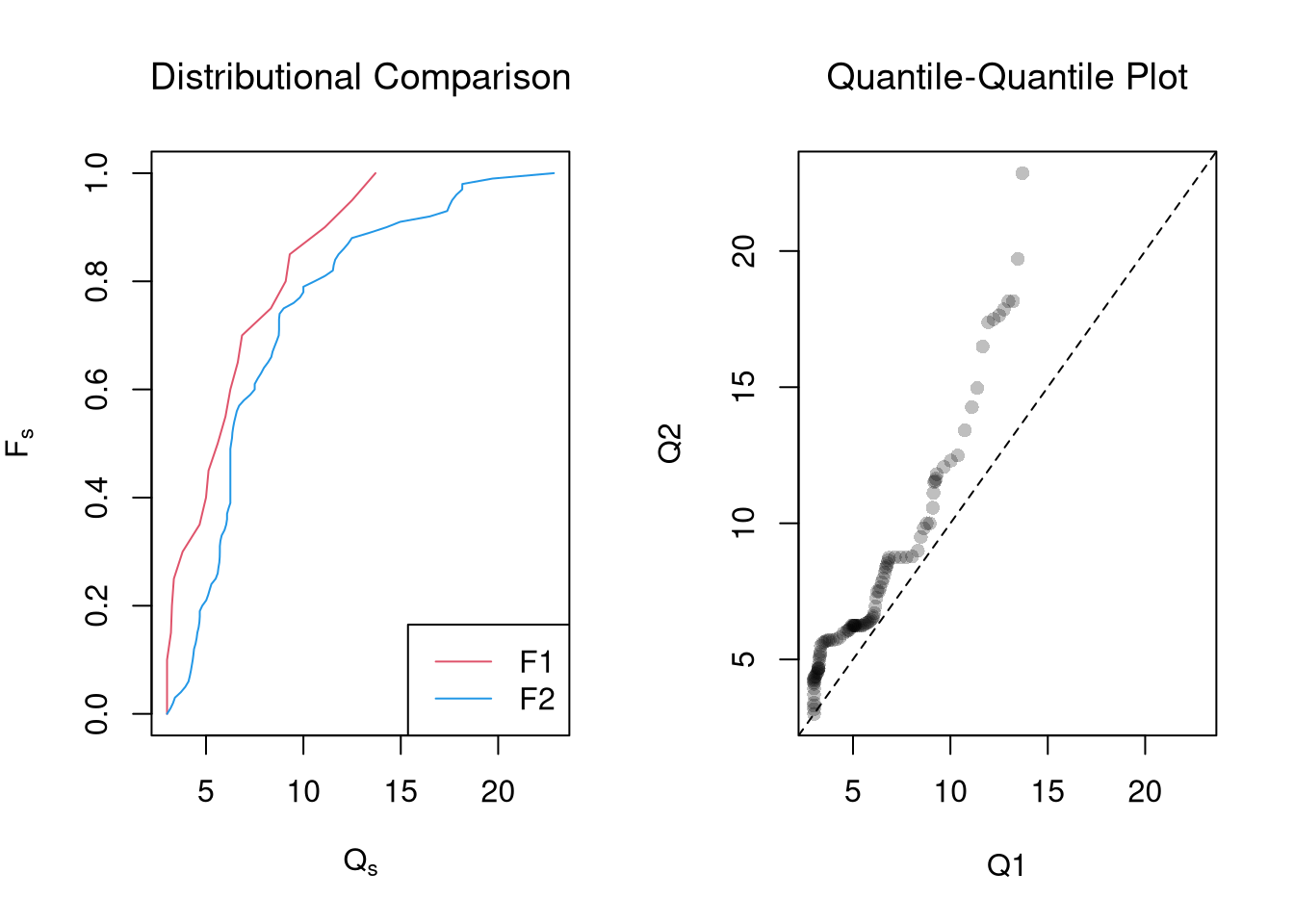
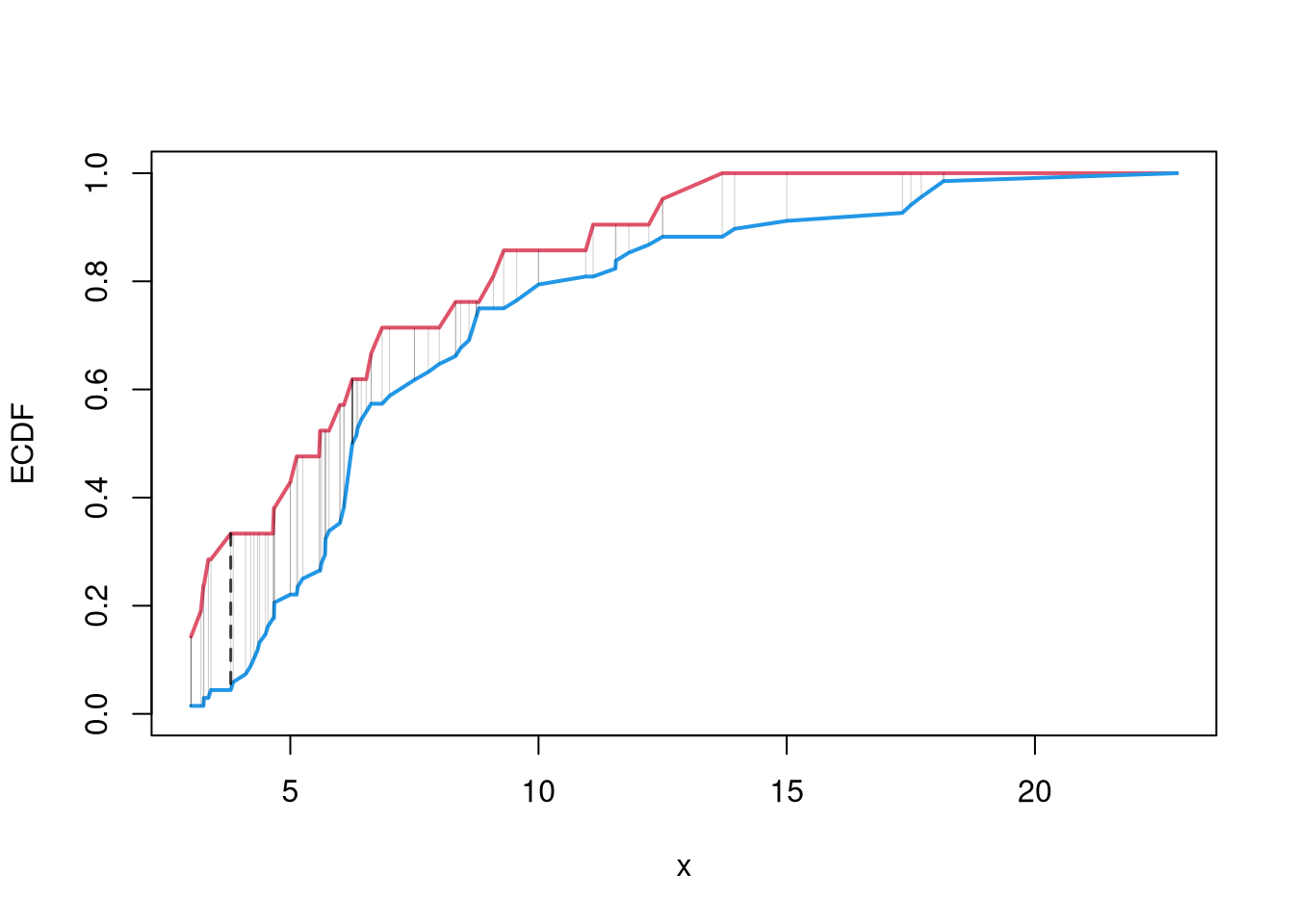
# Crossvalidated bandwidth for regression
xy_mat <- data.frame(y=CASchools[,'math'], x1=CASchools[,'income'])
library(np)
## Grid Search
BWS <- seq(1,10,length.out=20)
BWS_CV <- sapply(BWS, function(bw){
E_bw <- sapply(1:nrow(xy_mat), function(i){
llls <- npreg(y~x1, data=xy_mat[-i,],
bws=bw, regtype="ll",
ckertype='epanechnikov', bandwidth.compute=F)
pred_i <- predict(llls, newdata=xy_mat[i,])
e <- (pred_i- xy_mat[i,'y'])
return(e)
})
return( mean(E_bw^2) )
})
## Plot MSE
par(mfrow=c(1,2))
plot(BWS, BWS_CV, ylab='CV', pch=16,
xlab='bandwidth (h)',)
## Plot Resulting Predictions
bw <- BWS[which.min(BWS_CV)]
llls <- npreg(y~x1, data=xy_mat,
ckertype='epanechnikov',
bws=bw, regtype="ll")
plot(xy_mat[,'x'], predict(llls), pch=16, col=grey(0,.5),
xlab='X', ylab='Predictions')
abline(a=0,b=1, lty=2)
## Built in algorithmic Optimziation
llls2 <- npreg(y~x1, data=xy_mat, ckertype='epanechnikov', regtype="ll")
points(xy_mat[,'x'], predict(llls2), pch=2, col=rgb(1,0,0,.25))
## Add legend
add_legend <- function(...) {
opar <- par(fig=c(0, 1, 0, 1), oma=c(0, 0, 0, 0),
mar=c(0, 0, 0, 0), new=TRUE)
on.exit(par(opar))
plot(0, 0, type='n', bty='n', xaxt='n', yaxt='n')
legend(...)
}
add_legend('topright',
col=c(grey(0,.5),rgb(1,0,0,.25)),
pch=c(16,2),
bty='n', horiz=T,
legend=c('Grid Search', 'NP-algorithm'))